SmartTM Library
Description
Design of our SMART(TM) Library
ChemDiv offers a new “Smart” library of drug-like compounds for hit-finding screens. The Library has been assembled using ChemDiv’s in house structural biology insight, molecular simulation-modeling, virtual screening our novel chemistries and medicinal chemistry filtering/ranking of the resulting scaffolds. The design of the Smart library is based on our successful “Targeted Diversity” concept (Fig.1).
ChemDiv introduced the concept of Targeted Diversity for design of high quality libraries of drug-like molecules, selectably focused against different biological targets. This concept also uses the superposition of a highly diverse chemical space on a representative assortment of divergent families & sub-families of targets, signaling, and transport molecules. The Targeted Diversity concept is a broadly applicable platform approach, for it can be used to design and/or select high quality libraries of drug-like compounds focused in individual therapeutics areas, target families, or control pathways.
A major benefit of the Smart Library approach is that it represents a thoughtful and multi-purpose diversity set, incorporated it into one single compound library for different screening goals. Those goals may include:
- “Difficult” targets (no target / ligand structure(-s) known, study and interrogation of cellular processes (e.g. apoptosis, cell cycle, etc.);
- Signaling pathways (e.g. WNT, Hh, RTK, Ras, etc.) and “eclectic” biological targets, including cellular processes (e.g. apoptosis and cell cycle);
- Protein-protein interactions (the most «hot spots», e.g. XIAP, pGPCRs, beta-catenin, etc.);
- An entire selected therapeutic area that incorporates multiple biological targets;
As a starting point, the following main target families (encompassing 300+ various targets in total) were considered:
- GPCR target family;
- Ion channels (both ligand- and voltage-gated);
- Nuclear receptors;
- Enzymatic targets (kinases, phosphodiesterases, phosphatases, proteases, HDAC, sulphotransferases, esterases, COX, MAO, etc.);
- Transporters (serotonin transporters, dopamine transporters, amino-acid transporters);
- Effector proteins (pro- and anti-apoptotic, BCL2, Diablo, integrins, etc.);
- Non-human targets.
Each of these target families contains various orthogonal targets that were already annotated as interacting with small molecules. These small-molecule prototypes were used to select focused sets against each individual target from a given target family and further refined, thus forming the Smart-1 library.
The current generation of this library is based on approximately 500 small molecule sets. Each of these sets was focused against distinct biological targets that belong to different classes and sub-families of targets (list of targets of interest is shown). The selection process for these sets involved identifying active ligands/inhibitors as prototypes existing in the patent and research literature or databases and performing bioisosteric replacement (Fig.2) strategies on those (e.g. known peptide ligand may be substituted with small non-peptide peptidomimetic).
Subsequently, a similarity search based on these strategies was conducted within ChemDiv’s collection for possible augmentation of the rational set. Other techniques included computer-assisted 3-D pharmacophore matching. The directed synthesis of new chemotypes with functionality mimicking recognition elements (shapes, helix, turns, etc. Fig.3) of known active ligands/inhibitors was also performed. In some cases, proof of concept has been established with in-house biological data. A special effort has been made to select respective compounds and synthetic templates with good IP potential, as evidenced by Reaxys and SciFinder sub-structure searches. ChemDiv’s medchem filters (MCF) ensured high quality and drug-like properties of selected molecules.
Based on the defined “refinement” criteria a certain number of compounds from each “single target focused set” were selected, thus forming the final Smart-1 library
Such “refinement” criteria include, but not limited to:
- Available annotation data (literature, patent search, in-house research);
- Diversity of scaffolds and their further population with individual compounds;
- Bioisosteric replacement strategies, especially heterocyclic morphing;
- Computer-assisted 3-D pharmacophore matching;
- Special preference given to chemotypes with functionality mimicking recognition elements (shapes, helix, turns, etc.) of known active ligands/inhibitors for the considered targets;
- Special preference given to scaffolds with good IP potential (as evidenced by Reaxis and SciFinder sub-structure searches);
The current generation of the Smart-1 consists of more than 500 individual target-specific sub-sets (focused sub-libraries, 25 to 250 molecules each). It also encompasses around 1,900 diverse chemical templates and more 600 unique heterocycles. The total size of this library is 55,000 individual drug-like molecules
Related
Library Composition
ADVANTAGES
Smart-1 library may be a potential source of high-quality hits in screening against ‘difficult’ targets with limited or no structure/ligand information, as well as ‘eclectic’ biological targets, including cellular processes and signaling pathways
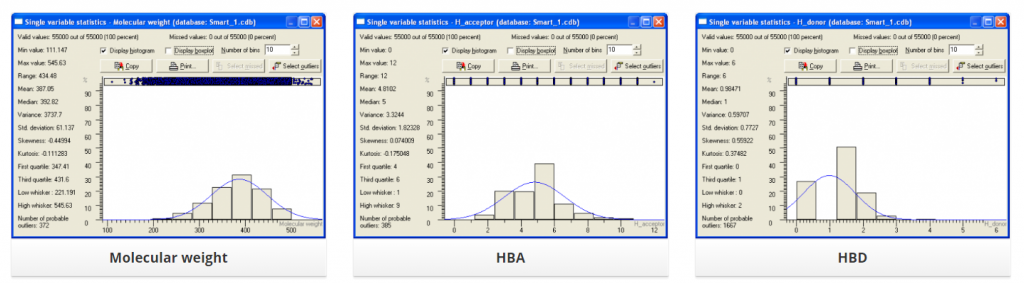
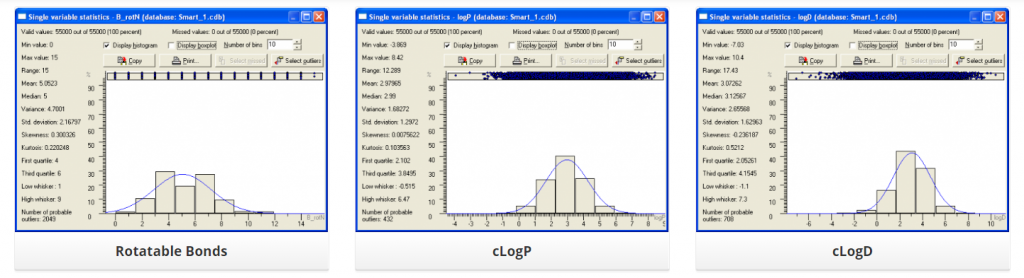
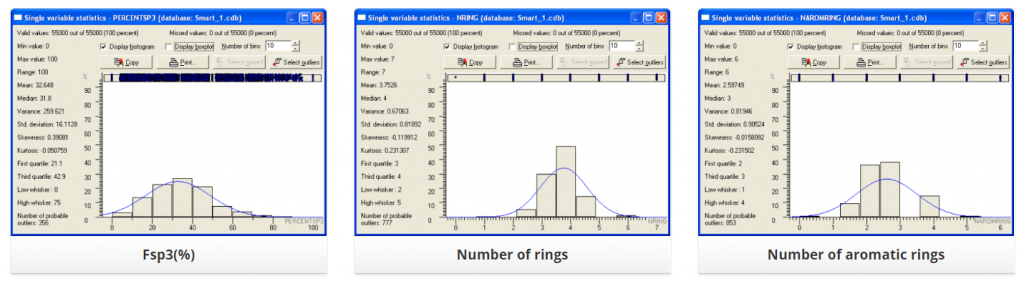
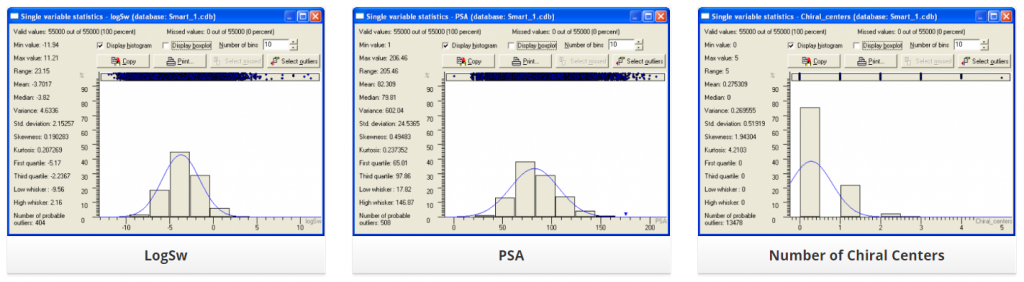
Distribution of several physic-chemical properties across this library is summarized below: